Asynchronous evolution on LSTM neural networks: Introducing PrAsE library
I was thinking about creating a small library for my colleagues, which they can use to learn about and further develop the idea of genetically programmed neural networks. I am kind of a fan of biological evolution and genetics, which can bring a lot of inspiration and learning about how not only organisms, but all of the world’s diversity and natural beauty of mild forests, flowering meadows and moss-covered rocks develop from the tiniest particles. And where else should we apply evolution than in artificial intelligence? To evaluate the concept, I created a simple LSTM neural network builder and passed it to the evolutionary algorithm. Let me first explain all these topics a bit more.
The long short-term memory network, or LSTM, is a very useful type of recurrent neural network, especially in the cyber security area, where you want to analyze and understand a long series of data with shorter or longer gaps inside them. Recurrent networks in general are suitable for analyzing long data rows, because they contain one or more hidden layers and utilize the feedback loops. In order to properly parametrize the hidden layers as well as the activation function, optimization etc., you need to have a good understanding of the problem you are solving. That is where genetic programming comes in.
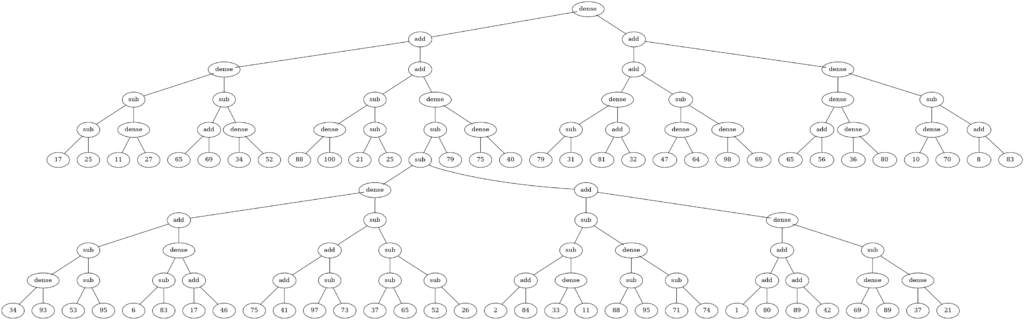
Genetic programming utilizes evolutionary algorithms to create programs. You can imagine pieces of source code like functions to be a kind of “genes”, that you want to mix and replicate to get the best result, that is a program solving a particular task. Here I am using the same strategy to add neural network layers with a parameter specifying the count of nodes. Take a look at the following picture, which illustrates the result of genetic programming. The algorithm builds a tree that represents an individual being from the perspective of an evolutionary algorithm. Each node in the tree is a gene, including the number constants below or operations such as unsigned add and subtraction, which I introduced here, and a function to add a dense layer to the neural network model. By compiling the tree, all methods are called, hence a model is built with the amount of dense layers specified in the tree.
The small library that I created and that tries to connect both of those worlds from the artificial intelligence area, that is genetic programming with neural networks, is available on GitHub under the name PrAsE: predictive asynchronous evolutionary builder. What I wanted was a really simple-to-use library and template that my colleagues can extend to validate their own concepts. The library is split into three types of objects: gods, prophets and spirits. Gods represent different parametrization of genetic programming and the evolutionary algorithm, prophets store the data and provide interface to their spirit, which is the actual neural network model representation. The prophets should observe the data stream in real time, that is the events that are being collected from security or other types of systems. Then, periodically on the asynchronous loop, they should get inspired by a god to build the model in their spirit. The asynchronous approach makes sure the data observation is not stopped during the long process of inspiration, which involves the actual run of the genetic programming and evaluation by training and testing the models.
Look at the following code, that should be triggered in the init time of the application:
|
1 2 3 4 5 6 7 8 9 10 |
# Evolutionary algorithm apollo = prase.God(generations_count=10) # Neural networks and data storage pythia = prase.TimeProphet( name="pythia", timestamp_field="timestamp", span=5, resolution=10 ) |
The only parametrization here to the god object named apollo is the generation count for the evolutionary algorithm. The time prophet object named pythia says how structured the time window for the observed data should be and which attribute of the incoming events to observe (timestamp field). The time window here is defined by the resolution, which is 10 seconds. That means we want to make predictions for the following ten seconds. Then, the span specifies the count of the resolutions that should be used for training the model for each one resolution output. Here it means that for each 10 seconds output there should be 50 seconds of input.
After the observation of the data stream via (pythia.observe(event)) is started, there should be a periodic call to build the model and make a prediction. See the following code:
|
1 2 3 4 5 6 7 8 9 10 11 12 |
# Transform the observed data into data suitable for the model print("Pythia is meditating.") pythia.meditate() # Create, train, test and save the neural network model using genetic programming print("Pythia is getting inspired by Apollo.") loop.run_until_complete( apollo.inspire(pythia) ) predicted_value = pythia.predict() |
The first call transforms the observed data to the actual time window and its training and testing datasets. After that, the long inspiration/evolution process is called on the asynchronous loop. There can be another, but more frequent periodic check, if pythia is ready (pythia.is_inspired()) and only after that the prediction can be called. Or the code for inspiration and predictions can be located in the same function, as specified in the snippet above.
This is just a brief introduction both to the theory of artificial intelligence concepts of neural networks and genetic programming, as well as to the small PrAsE library. The library is open-source, built on top of the wonderful DEAP library and TensorFlow. It automatically synchronizes the models into files and provides simple calls to get the visualizations of both the model and the genetic programming algorithm’s result. The library is available here: https://github.com/pagan-coder/PrAsE/
Please feel free to experiment and send me feedback!




